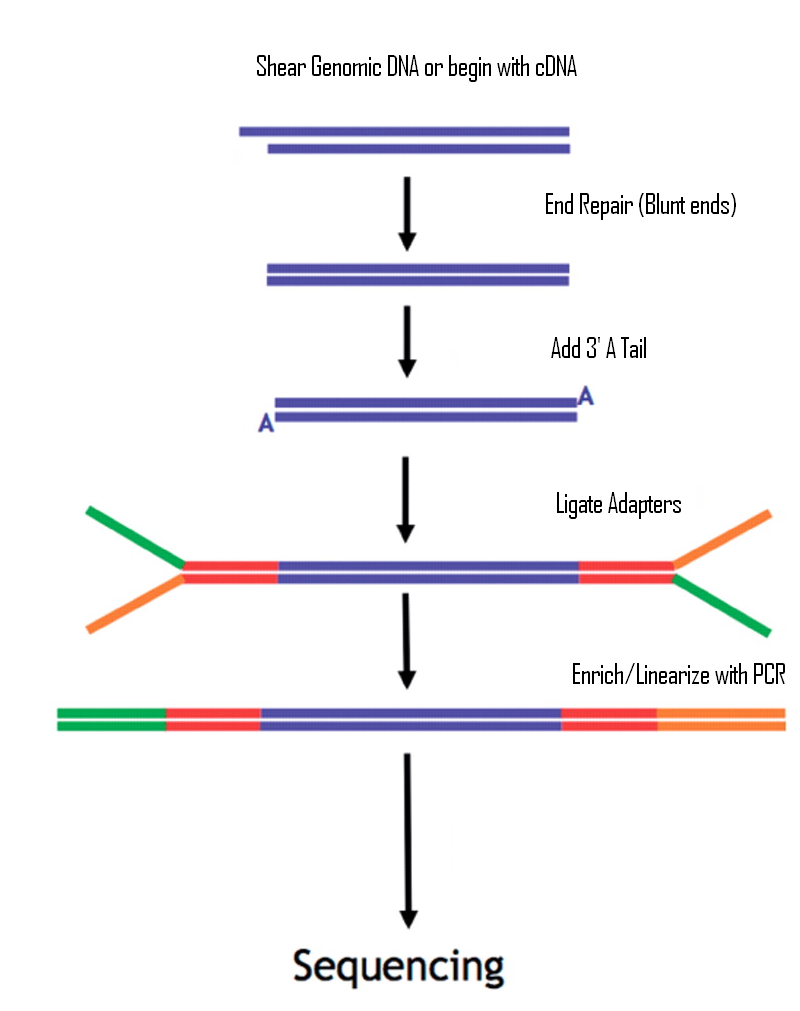
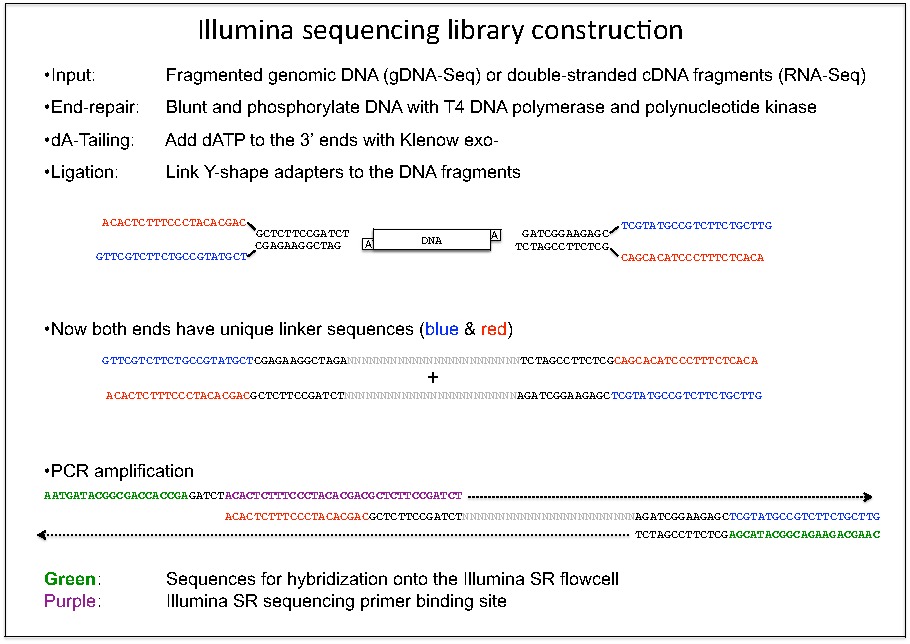
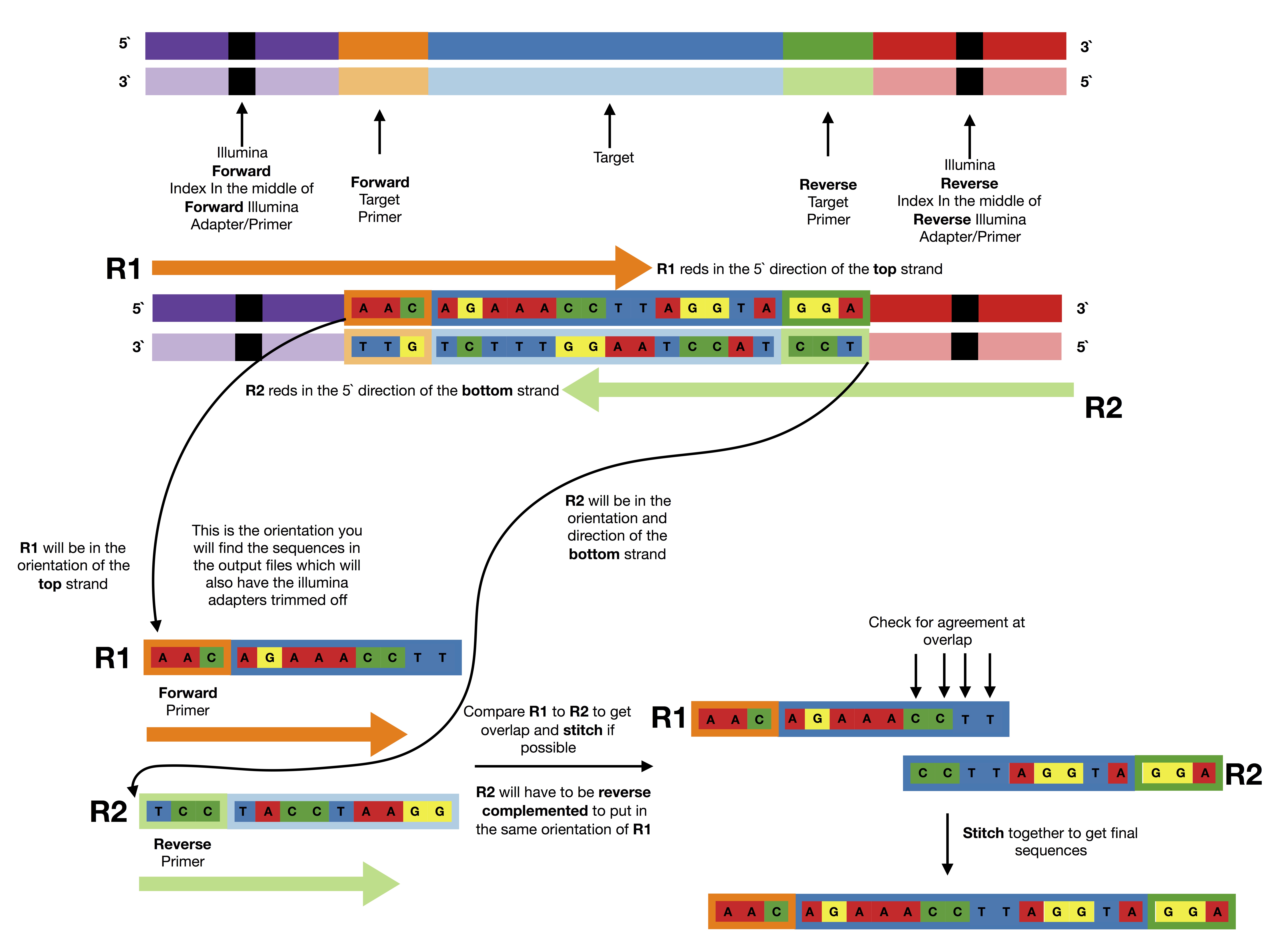
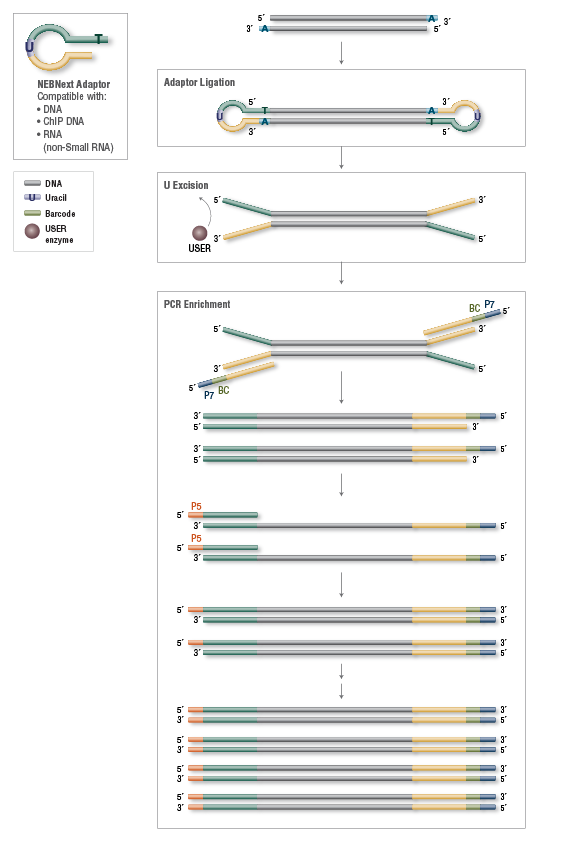
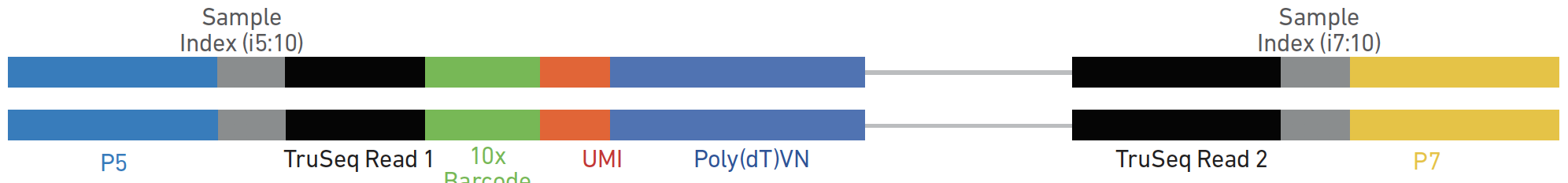
Dual indexed design of in-Drop single-cell RNA-seq libraries improves sequencing quality and throughput | bioRxiv

Reproducibility and repeatability of six high-throughput 16S rDNA sequencing protocols for microbiota profiling - ScienceDirect

Understanding the differences between Illumina index chemistries and anchor function - Illumina Knowledge

Overview of the different methods used for adapter addition to antibody... | Download Scientific Diagram

Incorporation of unique molecular identifiers (UMIs) in TruSeq adapters improves the accuracy of quantitative sequencing | RNA-Seq Blog

Figure 1 from Quantitative Bias in Illumina TruSeq and a Novel Post Amplification Barcoding Strategy for Multiplexed DNA and Small RNA Deep Sequencing | Semantic Scholar
![Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext) [PeerJ] Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext) [PeerJ]](https://dfzljdn9uc3pi.cloudfront.net/2019/7755/1/fig-1-full.png)
Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext) [PeerJ]